In order to share metabolomics analysis strategies and centralize tools and practices, the web-based platform Galaxy [1,2,3] is the core component of the W4M infrastructure. This web-based platform is open source with a very active community which insures up-to-date software releases and efficient support for both end-users and tools developers. Galaxy provides several interesting features for metabolomics tools integration compared to other workflow engines, which include: no known data size limitations [4], possibilities to automate pipelines, and to ensure reproducibility. Because of its web interface, this cross-platform system enables scientist without programming experience to design and run analysis workflows.
The main features of this platform are:
- a real benefit to users with results traceability and storage,
- the ability to share results between users/labs/platforms,
- the possibility to use a complete analysis workflow managing environment.
Galaxy provides an ergonomic interface for tools and workflows (Figure 1).
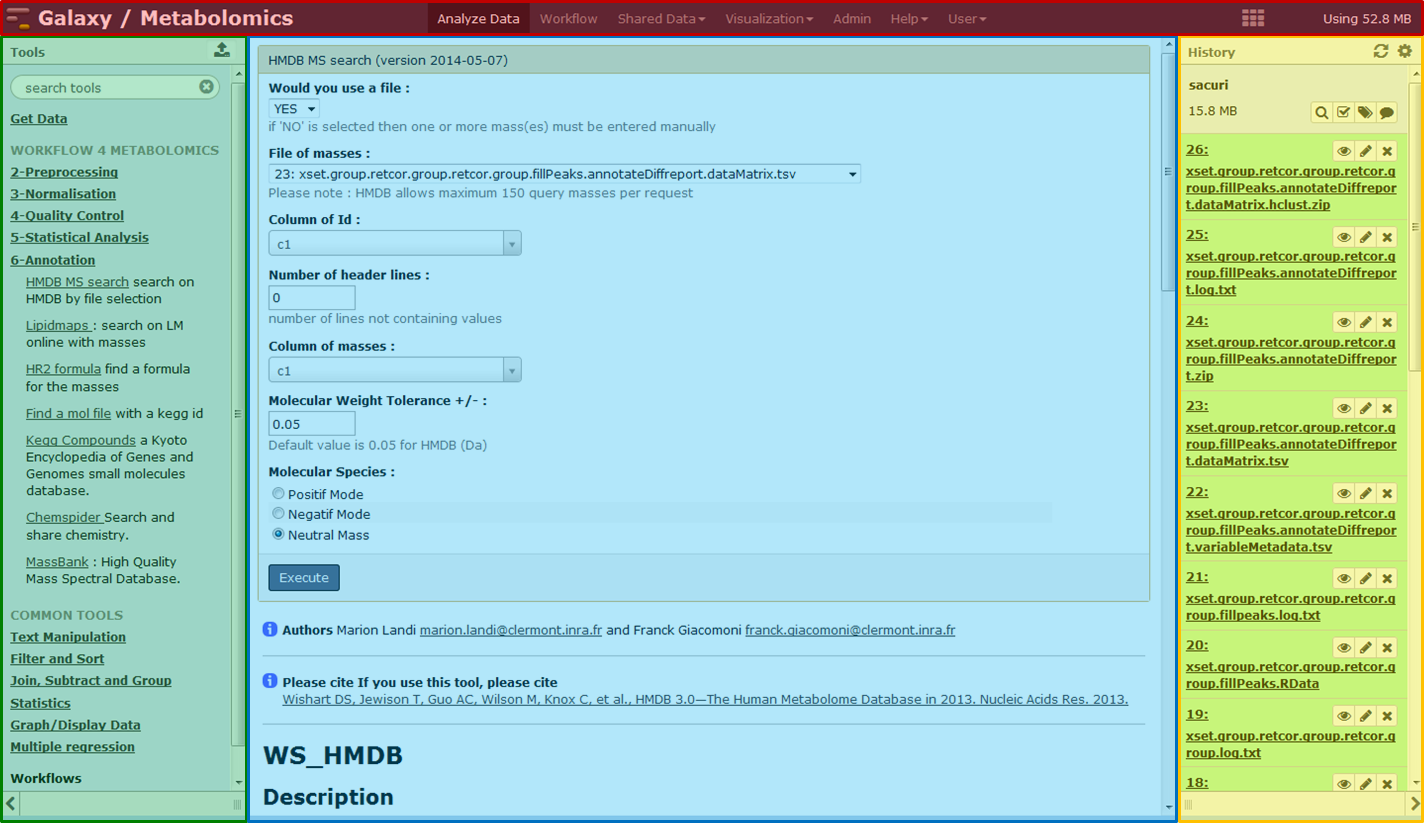
Figure 1: Main interface of the Galaxy interface. The main window is divided into a main menu (top red part), the tools list (from pre-processing to annotation, left green panel), the current history (right yellow panel) and the central visualization panel (in purple).
First, the interface is designed to be accessible: users without programming experience can easily specify parameters and run tools and workflows. Second, Galaxy is not only a computing tools runner but also an interesting way to share and publish analyses through the web and interactive web-based documents describing a complete analysis project.
In terms of computing infrastructure, the Galaxy platform “Workflow4metabolomics” is hosting on one of the French Bioinformatics Institute nodes: the Analysis and Bioinformatics for Marine Science (ABiMS) laboratory. Technically, Galaxy was deployed in a virtual environment based on VMWARE. Optimization efforts, such as connections pool or web services decoupling, allow a good level of scalability. Computing resources connection uses the standard DRMAA API, and is completed with a dedicated connector (tool runner) in order to make available adequate resources both in terms of high computing performance and memory amount (up to 1TB RAM). Finally, a shared and secure storage space completes this layer, essential to smoothly working treatments.