Workflow4Metabolomics Object Identifier: W4M00004
Digital Object Identifier: 10.15454/1.4811272313071519E12
Creator of the history: Yann Guitton & Sophie Goulitquer
Maintainer: Yann Guitton (yann.guitton at oniris-nantes.fr)
Creation|Updating date: 2016-12-01
Format: Workflow4Metabolomics Galaxy histories
W4M History: https://workflow4metabolomics.usegalaxy.fr/histories/list_published
Size: 260 Mo
Keywords: ectocarpus, algae, gcms, preprocessing, statistics and annotation
Description:
| Study: Characterization of the physiological variations of the metabolome in algae exposed to 3 different abiotic stress (salt concentration). | 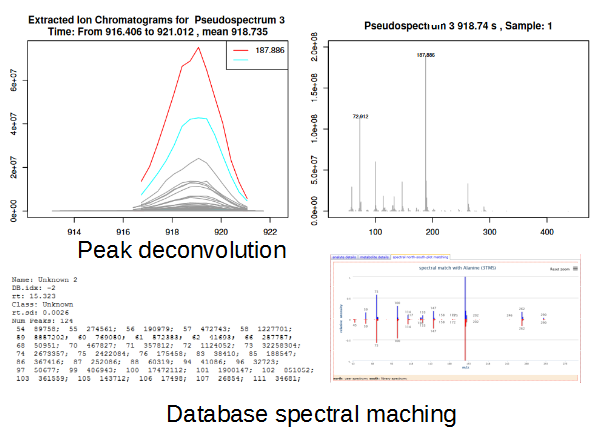 |
|
Dataset: The dataset contains 12 mzXML files created from Agilent RAW GC-MS file. 4 replicats per conditions: i) Condition 01: Fresh water strain in 1.6 ppt NaCl; ii) Condition 02: Fresh water strain in 32 ppt NaCl; iii) Condition 03: Sea water strain in 32 ppt NaCl |
|
|
Workflow: The workflow consists of the following steps: preprocessing with metaMS (runGC function) then Golm Metabolom Databases online search for annotation. The peakspectra.msp file can be exported and search locally against NIST database with MSsearch software (see "how to" section on W4M portal). The dataMatrix generated by metaMS is used for PCA analysis. |
|
|
Comments: Please note that, the original publication of the study by Dittami et al do not include the presented GCMS data or their multivariate interpretation. |
|
Rights: Creative Commons
Citations:
- Dittami,S.M. et al. (2012) Towards deciphethring dynamic changes and evolutionary mechanisms involved in the adaptation to low salinities in Ectocarpus (brown algae): Adaptation to low salinities in Ectocarpus. The Plant Journal, DOI: 10.1111/j.1365-313X.2012.04982.x
Raw data repository:
- N/A
Please find more referenced W4M histories here.