Workflow4Metabolomics Object Identifier: W4M00003
Digital Object Identifier: 10.15454/1.4811165052113186E12
Creator of the history: Etienne Thévenot and Philippe Rinaudo
Maintainer: Etienne Thévenot (etienne.thevenot at cea.fr)
Creation|Updating date: 2015-12-21
Format: Workflow4Metabolomics Galaxy histories
W4M History: https://workflow4metabolomics.usegalaxy.fr/histories/list_published
Size: 11 Mo
Keywords: diabetes, homosapiens, plasma, lcms, statistics
Description:
| Study: Identification of metabolomics signatures for patient stratification between type 1 versus type 2 diabetes mellitus. | 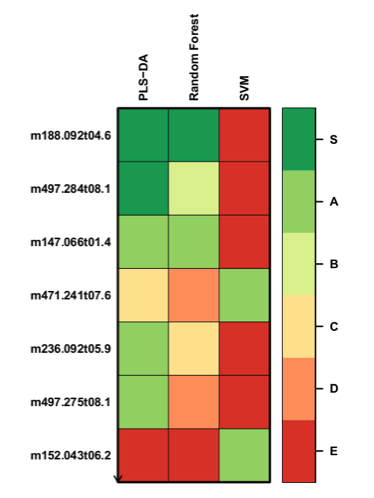 |
| Dataset: plasma samples from 69 diabetic patients were analyzed by reversed phase (C18) ultra-high performance liquid chromatography (UPLC) coupled to high-resolution mass spectrometry (Orbitrap Exactive). Age and body mass index (BMI) of patients are also provided as sample metadata since type 2 patients from this cohort are significantly older and with a higher BMI. The peak table contains 5,501 features (whose m/z and retention time have been matched against an in-house database). The intensities from the peak table have been log10 transformed. | |
| Workflow: The workflow consists of: PCA visualization (with the two diabetic types coloured on the score plot), univariate Wilcoxon hypothesis testing of median differences between diabetic types, OPLS-DA modeling of the diabetic type response, and selection of the metabolite significant signature for gender classification by PLS-DA, Random Forest or SVM. | |
|
Comments: The ‘diaplasma’ data set is also available in the biosigner R package from the Bioconductor repository. |
|
Rights: Creative Commons
Citations:
- Rinaudo P., Boudah S., Junot C. and Thevenot E.A. (2016). biosigner: a new method for the discovery of significant molecular signatures from omics data. Frontiers in Molecular Biosciences, 3. DOI:10.3389/fmolb.2016.00026
Raw data repository:
- N/A
Please find more referenced W4M histories here.