W4M is available on: here
Create a free account here.
Workflow4experimenters (W4E) course 2024 : Thanks for all participants !
Keep updated with our newsletter : SUBSCRIBE
For any questions or support: community.france-bioinformatique.fr/c/workflow4metabolomics/
Archives : 2016 W4E course at CNRS, Roscoff, 2017 W4E course at Pasteur, Paris, 2018 W4E course at Pasteur, Paris, 2020 W4E course at ULB, Brussels, 2021 Toulouse, 2023 Paris.
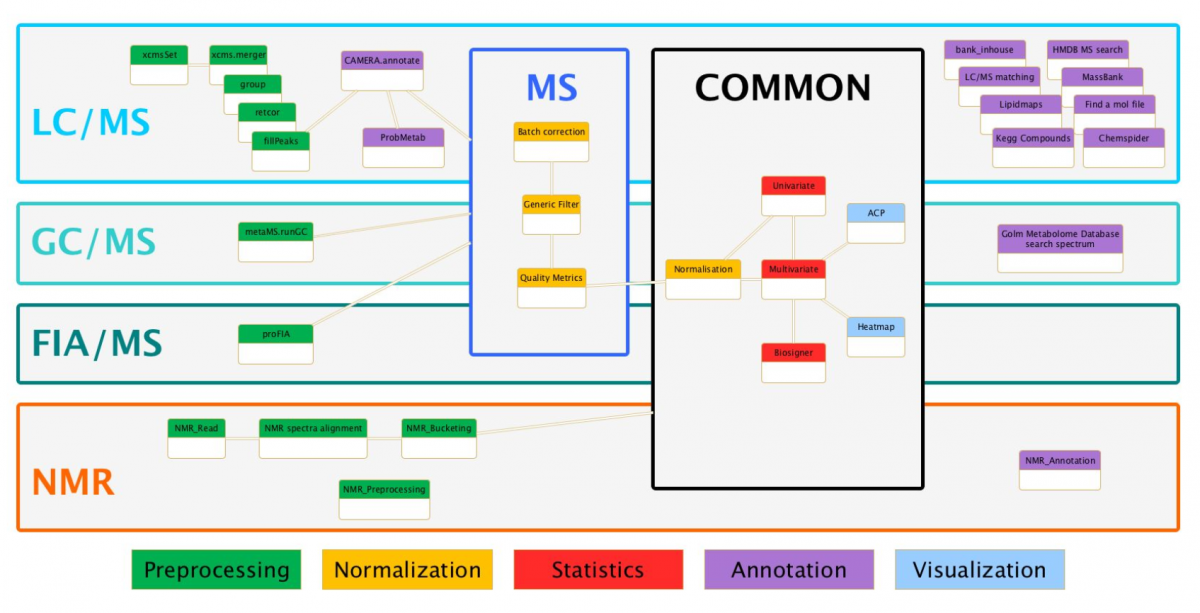
In the context of collaboration between metabolomics (MetaboHUB French infrastructure) and bioinformatics platforms (IFB: Institut Français de Bioinformatique), we have developed full LC/MS, FIA-MS, GC/MS and NMR pipelines using Galaxy framework for data analysis including preprocessing, normalization, quality control, statistical analysis (Univariate, Multivariate PLS/OPLS) and annotation steps. Those modular and extensible workflows are composed with existing components (i.e XCMS and CAMERA packages) but also a whole suite of complementary homemade tools. This implementation is accessible through a web interface, which guarantees the parameters completeness. The advanced features of Galaxy have made possible the integration of components from different sources and of different types. Thus, an extensible Virtual Research Environment (VRE) is offered to metabolomics communities (platforms, end users, etc.), and enables preconfigured workflows sharing for new users, but also experts in the field.
Publications :
Giacomoni F., Le Corguillé G., Monsoor M., Landi M., Pericard P., Pétéra M., Duperier C., Tremblay-Franco M., Martin J.-F., Jacob D., Goulitquer S., Thévenot E.A. and Caron C. (2014). Workflow4Metabolomics: A collaborative research infrastructure for computational metabolomics. Bioinformatics, http://dx.doi.org/10.1093/bioinformatics/btu813
Guitton Y., Tremblay-Franco M., Le Corguillé G., Martin J.F., Pétéra M., Roger-Mele P., Delabrière A., Goulitquer S., Monsoor M., Duperier C., Canlet C., Servien R., Tardivel P., Caron C., Giacomoni F., Thévenot E.A., Create, run, share, publish, and reference your LC–MS, FIA–MS, GC–MS, and NMR data analysis workflows with the Workflow4Metabolomics 3.0 Galaxy online infrastructure for metabolomics, The International Journal of Biochemistry & Cell Biology, 2017, ISSN 1357-2725, http://dx.doi.org/10.1016/j.biocel.2017.07.002. This paper is also available on the open archive HAL.
W4M is supported by the national infrastructure of metabolomics and fluxomics: MetaboHUB [MetaboHUB-ANR-11-INBS-0010, 2011] and the French Institute of Bioinformatics: IFB [ANR-11-INBS-0013236]
We thank genotoul bioinformatics platform for computing/storage resources and webhmia team for web portal hosting.